Optimizing RAG to Search 36 Million PubMed Articles in Milliseconds Instead of Minutes
June 28, 2024
About a week or so ago, I came across this pubmed-vectors repo on Twitter. The author, Kyunghyun Cho, had made several scripts that did the following:
- Downloaded PubMed data and saved it to a SQLite database. This resulted in a 36M row table and a database size of ~34GB.
- Generated embeddings based on the PubMed article titles and/or abstracts using Nomic AI's
nomic-ai/nomic-embed-text-v1.5model.
The final embeddings file is ~110GB in parquet format, where each embedding is a 768-element float32 vector.
I was intrigued by the part that mentioned it took several minutes for a single query, and I thought this would be a wonderful real-world use case for the experimental work I had previously done on state-of-the-art exact search with binary embeddings. So, I asked for the embeddings.
By leveraging previous work, how much quicker could we make the exact search?
Less than 200 milliseconds for an end-to-end response!
On my machine, an Intel CPU with 6 cores and 12 threads, I get < 100ms for a call to the RAG endpoint itself.
So, how did we do it?
Processing Data
The first part is processing the 110GB parquet file into 2.5GB of binary data.
- Use MRL to reduce the embeddings to 512 elements float32 elements, resulting in a 33% reduction in size.
- Convert the 512-element float32 vectors into 64-element uint8 vectors, representing 512 binary values.
Making the most of the Hardware
Now we have ~2.2GB of binary embeddings and ~275MB of IDs (36M int64 values).
The massive speedup comes from replacing the 768-element float32 vector with an 8-element uint64 vector (64-element uint8 vector reinterpreted). This enables comparing two vectors in a single SIMD operation:
julia> @code_llvm hamming_distance(q1, q2) ; Function Signature: hamming_distance(StaticArraysCore.SArray{Tuple{8}, UInt64, 1, 8}, StaticArraysCore.SArray{Tuple{8}, UInt64, 1, 8}) ; @ /Users/lunaticd/code/tiny-binary-rag/rag.jl:28 within `hamming_distance` define i64 @julia_hamming_distance_12257(ptr nocapture noundef nonnull readonly align 8 dereferenceable(64) %"x1::SArray", ptr nocapture noundef nonnull readonly align 8 dereferenceable(64) %"x2::SArray") #0 { top: ; @ /Users/lunaticd/code/tiny-binary-rag/rag.jl:33 within `hamming_distance` ; ┌ @ simdloop.jl:77 within `macro expansion` @ /Users/lunaticd/code/tiny-binary-rag/rag.jl:34 ; │┌ @ /Users/lunaticd/code/tiny-binary-rag/rag.jl:14 within `hamming_distance` ; ││┌ @ int.jl:373 within `xor` %0 = load <8 x i64>, ptr %"x1::SArray", align 8 %1 = load <8 x i64>, ptr %"x2::SArray", align 8 %2 = xor <8 x i64> %1, %0 ; ││└ ; ││┌ @ int.jl:415 within `count_ones` %3 = call <8 x i64> @llvm.ctpop.v8i64(<8 x i64> %2) ; │└└ ; │┌ @ int.jl:87 within `+` %4 = call i64 @llvm.vector.reduce.add.v8i64(<8 x i64> %3) ; └└ ; @ /Users/lunaticd/code/tiny-binary-rag/rag.jl:36 within `hamming_distance`
Additionally, comparing one element against every other element in the list falls under "embarrassingly" parallel algorithms. Assuming we have multiple cores, each core can process its own chunk of the list, and then the results are aggregated at the end.
If we have 1000 elements, 4 cores, and we want to find the top 5 closest pairs, each core would process 250 elements and keep its own heap with a max capacity of 5 elements. Then we wait for all 4 cores to return their results and do a final 20-element sort, returning the top 5 of the 20 results.
@inline function hamming_distance(x1::T, x2::T)::Int where {T<:Integer} return Int(count_ones(x1 ⊻ x2)) end @inline function hamming_distance( x1::AbstractArray{T}, x2::AbstractArray{T}, )::Int where {T<:Integer} s = 0 @inbounds @simd for i in eachindex(x1, x2) s += hamming_distance(x1[i], x2[i]) end s end function _k_closest( db::AbstractMatrix{T}, query::AbstractVector{T}, k::Int; startind::Int = 1,
Here's the complete code for the RAG service.
Embedding Text
Luckily, embedding with just a CPU is not a bottleneck, and it, in fact, runs faster than the RAG search, at around 20-30ms.
The code for embedding is taken from Nomic AI's Hugging Face model page and the portion to binarize the embedding is copied over from sentence-transformers:
quantized_embeddings = np.packbits(embeddings > 0)
This is hooked up to a FastAPI POST endpoint that returns a 64-element uint8 vector.
from fastapi import FastAPI, HTTPException from pydantic import BaseModel import numpy as np import torch from transformers import AutoTokenizer, AutoModel import torch.nn.functional as F import time import uvicorn import fire MATRYOSHKA_DIM = 512 app = FastAPI() tokenizer = None model = None class TextInput(BaseModel): text: str
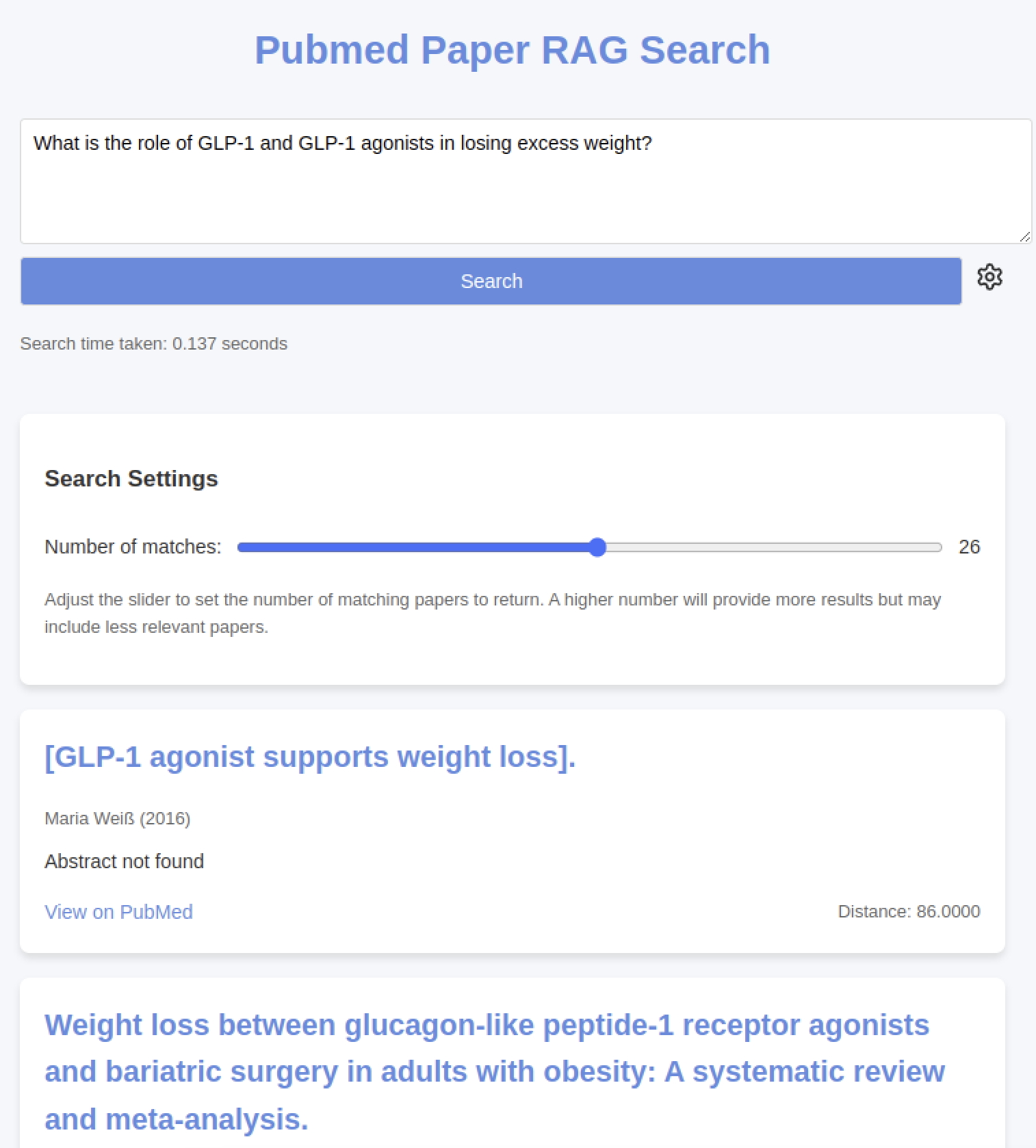
The App
There are many ways to set up this part. I wanted something simple. A Go app with an HTML template using plain CSS and JavaScript does everything we need to do, so why not? We can even ship this whole part as a binary.
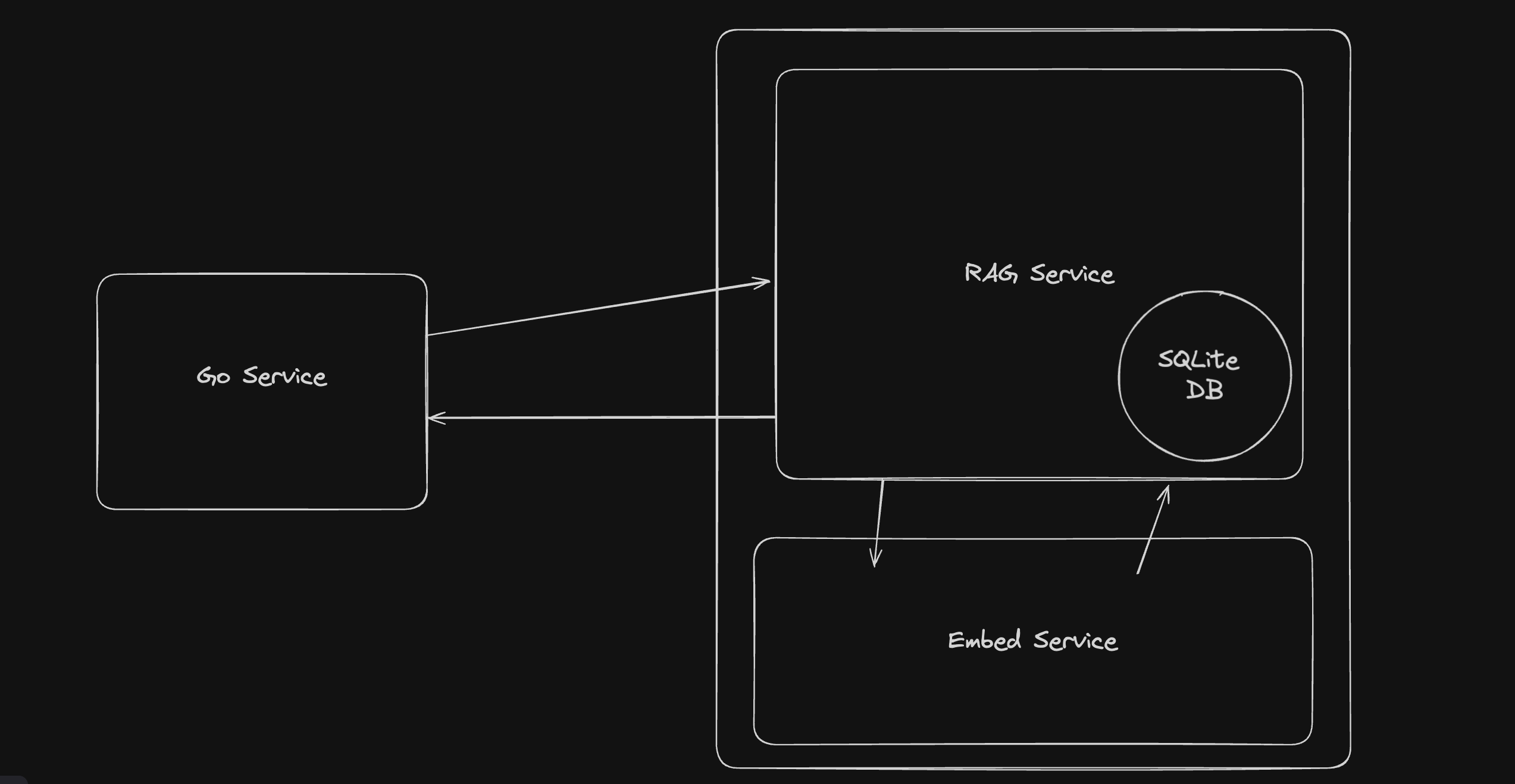

So that's it - enter some text, hit search, search PubMed with a RAG, and a couple hundred milliseconds later, you have the most relevant papers for your search.